

State of the art: RNA splicing in Severe ALD
There has been little progress in treatment of alcohol induced cirrhosis or hepatitis beyond abstinence and supportive care. This post looks at a recent study investigating the molecular biology underlying cases of persistent severe ALD.
Acute alcohol hepatitis and alcohol cirrhosis affects about 3 million individuals worldwide each year. Treatment consists of supportive care, correction of electrolyte and other imbalances, and abstinence from alcohol. Acute cases may or may not respond to steroids. Steroid treatment is discontinued if non responsive due to increased infection risk.
In 10-20% of cases the disease will continue to progress despite discontinuance of alcohol use. There are no specific treatments available.

This study was published in the journal Nature Communications September 10, 2025. The investigators utilized a number of techniques analyzing specific proteins, transcription factors, and RNA from liver tissue. Tissue samples were obtained from explanted livers from patients with severe alcohol hepatitis or alcohol cirrhosis. Control samples were taken from healthy tissues resected for unrelated reasons.
The investigators sought to identify specific bio markers and mechanisms responsible for disease progression and failure of regeneration and homeostasis. The study looked at the cellular milieu, deep RNA sequencing at the single cell level, and post transcriptional modifications.
The following section is a brief review of molecular biology in order to provide a framework for the remainder of the discussion. (green background)
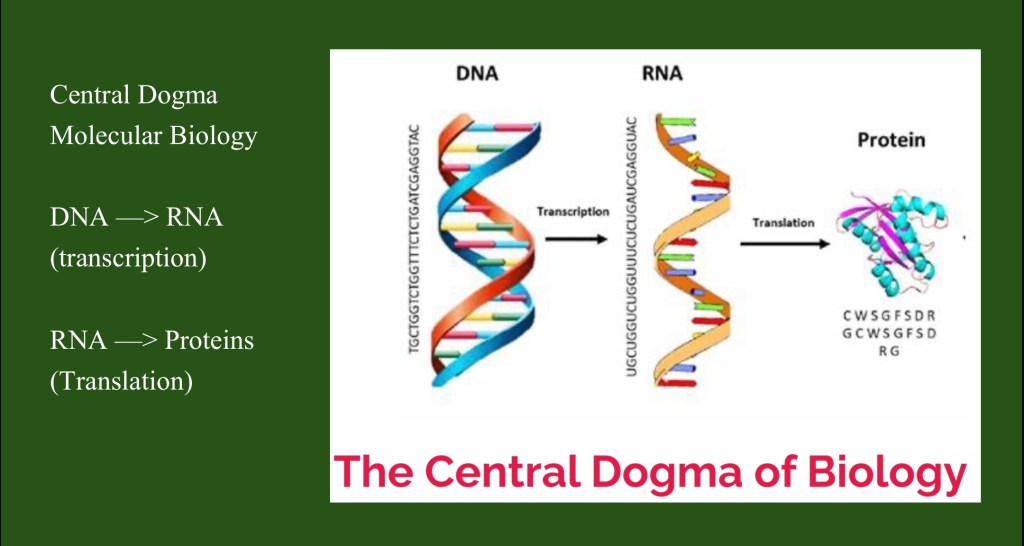
The central dogma of molecular biology was first proposed by Francis Crick. It states that in living things genetic information flows in one direction. DNA code is transcribed to RNA. RNA is translated into proteins.
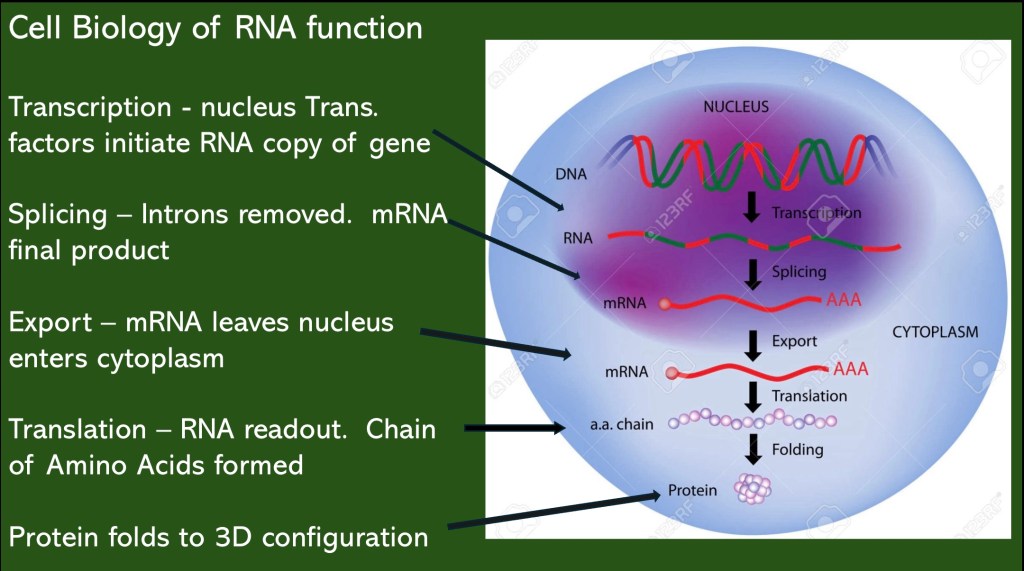
More detailed look into the processes involved in transcription and translation.
DNA is located in the nucleus. When a gene is activated it is transcribed into a single stranded RNA copy. There are additional modifications necessary prior to translation into a protein.
The initial RNA copy contains segments called introns and exons. RNA binding proteins orchestrate removal/splicing of introns. The remaining exons will form the final messenger RNA (mRNA).
The mRNA is exported from the nucleus and enters the cell cytoplasm. There it will attach to a ribosome and initiate formation of an amino acid chain which will fold into a protein.
This means that any proteins regulating gene expression or RNA binding will need to enter the nucleus where DNA is located. This process requires the ability to translocate through the membrane.
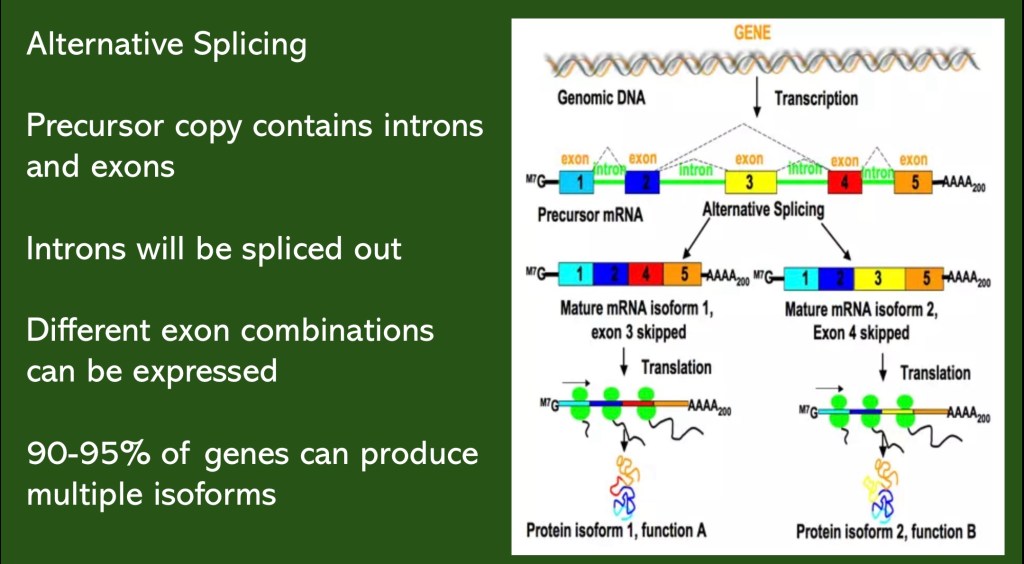
There are only about 20,000 coding genes in the human genome yet there are at least 100,000 proteins. The discrepancy is resolved by the ability of a given gene to produce more than one protein through RNA alternative splicing.
In the schematic above the initial RNA copy contains 5 exons. These can be rearranged to form 2 mRNAs each containing 4 exons. They can each code for proteins with differing properties. These related proteins are known as isoforms.
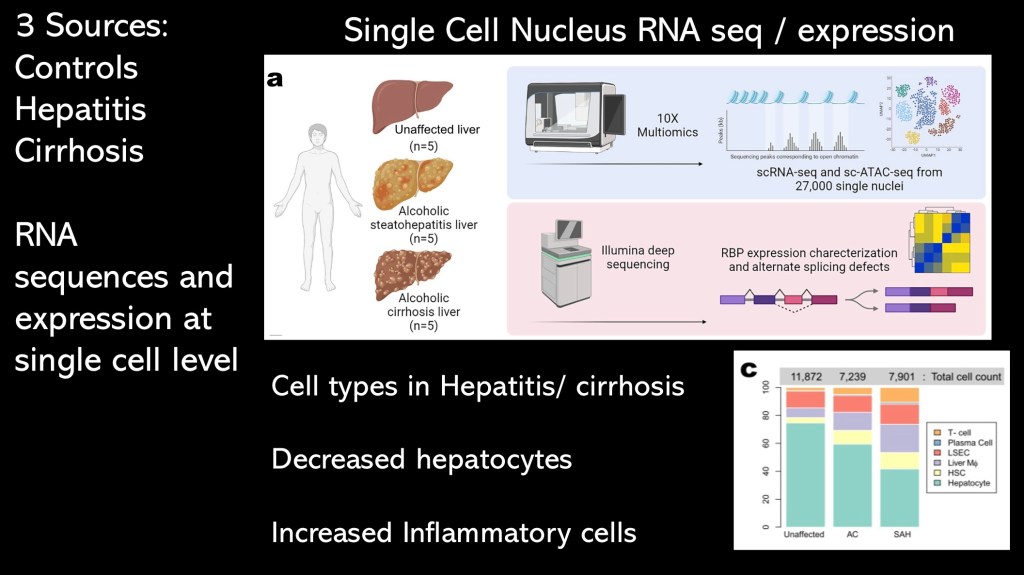
This study utilized technology capable of characterizing RNA at the single cell level. This allows for identifying local conditions and effects which may be obscured in large batch analysis.
The bar graph shows the relative tissue composition in healthy, hepatitis, and cirrhosis tissue.
Compared to controls the disease tissue contained fewer hepatocytes and greater numbers of inflammatory cells such as macrophages and T-lymphocytes. Within this inflammatory environment transcription errors and changes in chemical signaling are more likely to occur.
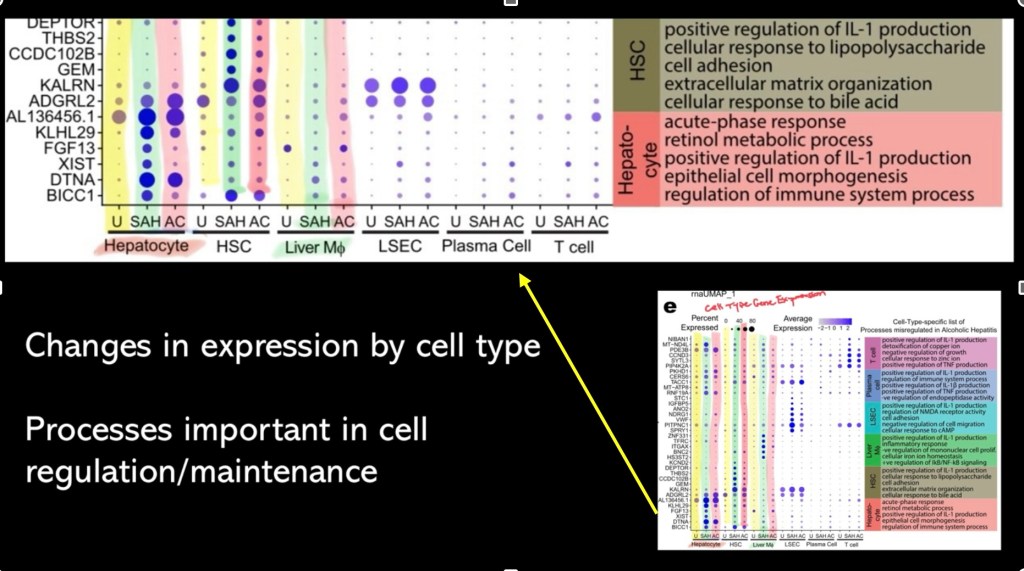
Plot showing expression of genes important in alcohol liver disease by cell type. The corresponding function is shown on the right.
The topmost image is a closeup of the chart showing findings for hepatocytes and hepatic stellate cells
Added color coding shows column correspond to:
U – unaffected controls (yellow)
SAH – severe alcohol hepatitis (green)
AC – alcohol hepatitis (red)
Blue circles represent concentration of the protein present. Reading across the rows allows evaluation of changes, increased or decreased expression, occurring in acute disease for each cell type.
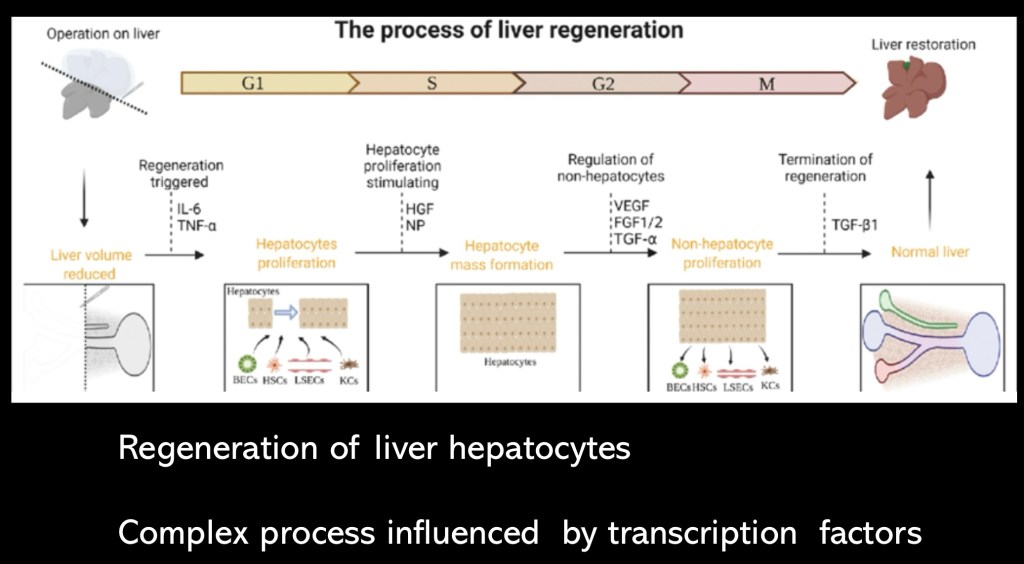
Hepatocytes are endodermal in origin They are related to the cells lining the intestine. In order to regenerate functional liver cells a progression of events must occur. Specific factors and co factors must be present to induce change in hepatocytes to proliferative cells capable of division and formation of new cells. This requires activation of quiescent genes which were active during early life and development.
Following this a switch to activate and transcribe genes required for transition to mature functional hepatocytes needs to occur. The central hypothesis in this study posits that dysfunctional alternative splicing (AS) is a key mechanism in cases of disease progression despite abstinence and steroid treatment.
The next section describes the Western Blot. This is a technique used to characterize proteins in a tissue sample or culture.
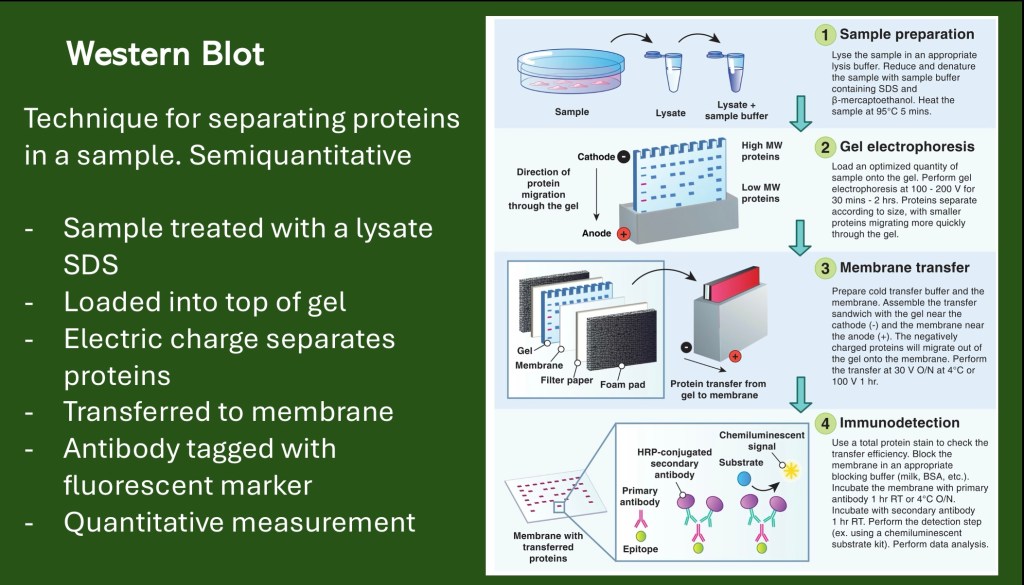
The western blot is a means of separating proteins in a sample based on size.
· The sample is treated with a lysate to break open the cells. A detergent, typically SDS is added. This surrounds the proteins with negative charge.
· The sample(s) are loaded into wells at the top of a gel.
· An electrical current is applied to the gel.
· The charged proteins move through the gel. Smaller molecules move faster than larger ones.
· The sample is transferred to a membrane along with a marker allowing for quantitative analysis
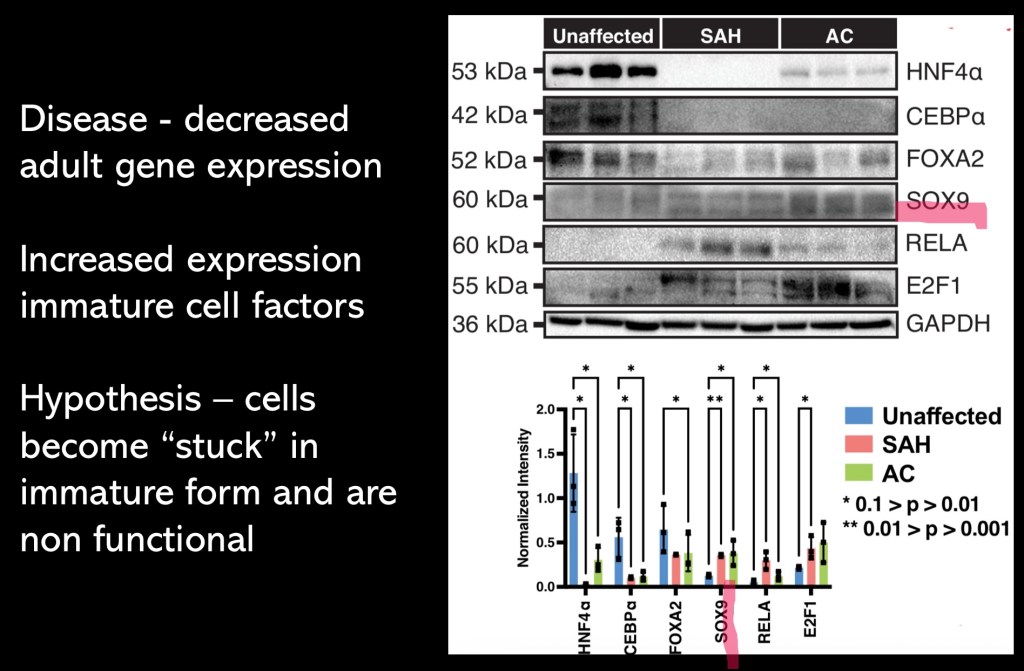
Western blot and graph showing proteins expressed in SAH and AC liver samples vs controls. The first three columns on the left in the graph represent proteins critical in maintenance of mature hepatocytes. On the right the proteins are primarily involved in early development and differentiation.
For example HNF 4 α (hepatocyte nuclear factor)is a transcription factor. It operates in the nucleus controlling genes involving glucose and fatty acid metabolism. It has a developmental role in cellular maturation.
SOX 9 is also a transcription factor. It is involved in early development of the liver, pancreas, and skeletal systems.
The pattern is consistent with under expression of genes important in maintenance of mature hepatocytes and over expression of genes involved in early development. This supports the hypothesis that underdevelopment of regenerative hepatocytes is a central abnormality in these cases.
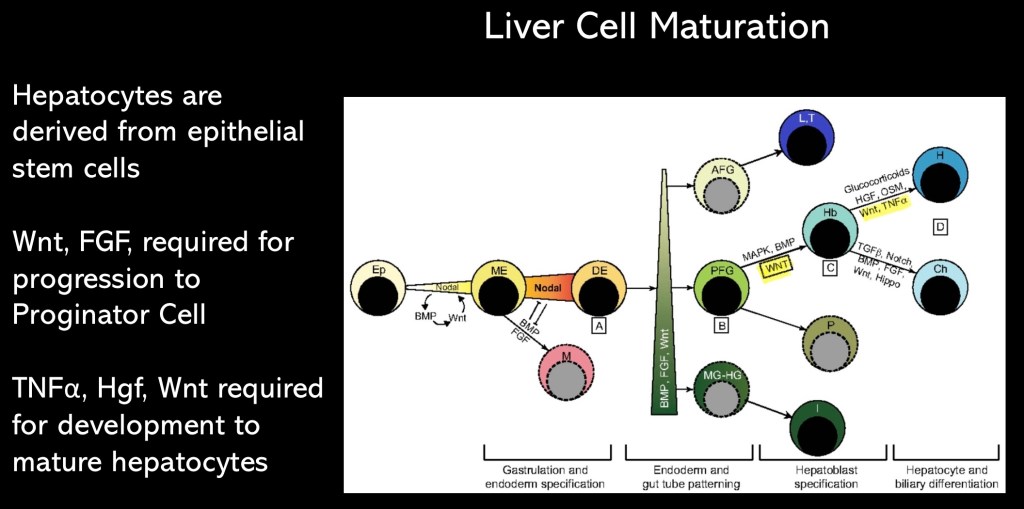
Flow chart of stages in maturation of hepatocytes from early endoderm to maturity. Note that the factors WNT, and TNFα are required to develop into functional hepatocytes.
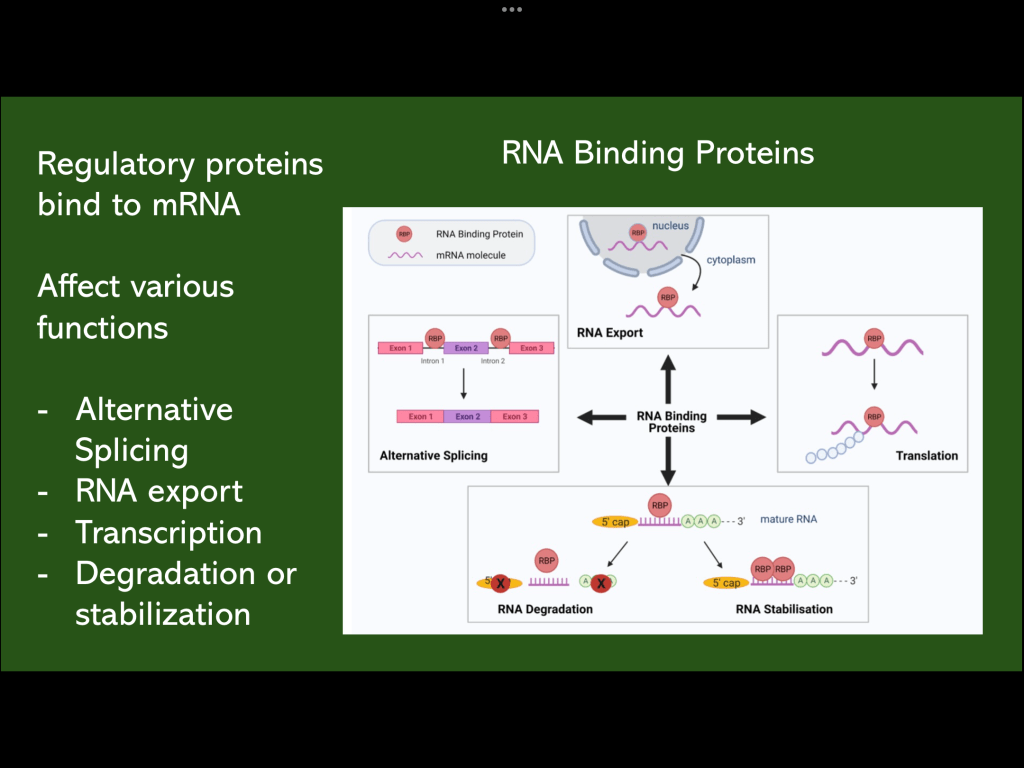
The next part of the study focuses on RNA splicing and RNA binding proteins. RNA binding proteins function in mRNA transport from the nucleus, alternative splicing, protein translation and RNA degradation following translation.
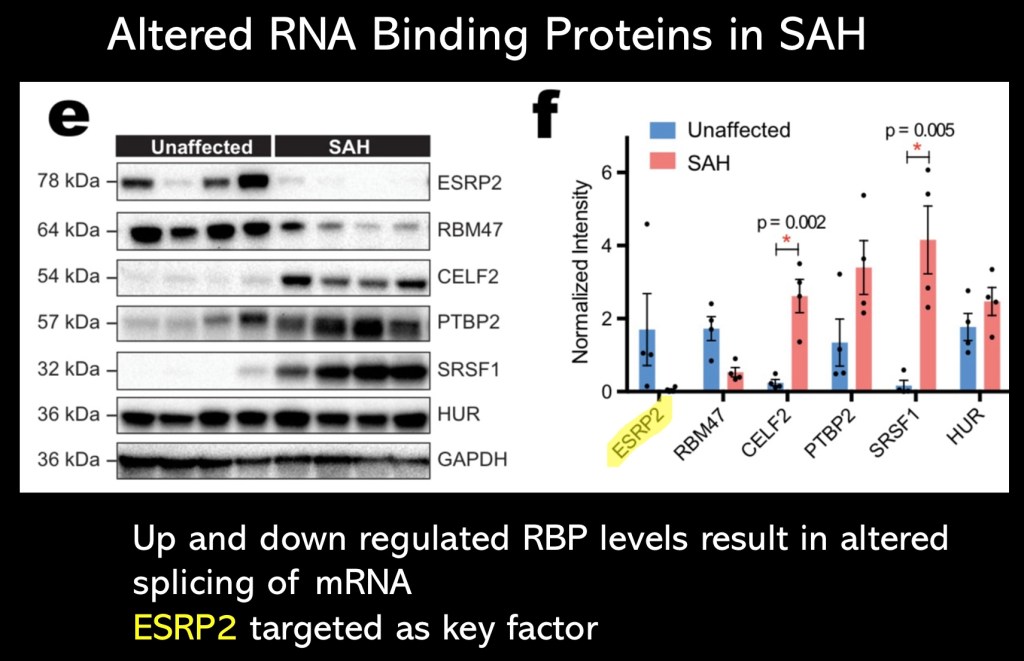
Level of RNA binding protein expression in controls vs SAH. The authors focused on marked decrease in level of ESRP2. It is known to be essential in regulation of pathways essential in maintaining mature hepatocytes. 20% of alternative splicing is controlled by ESRP2.
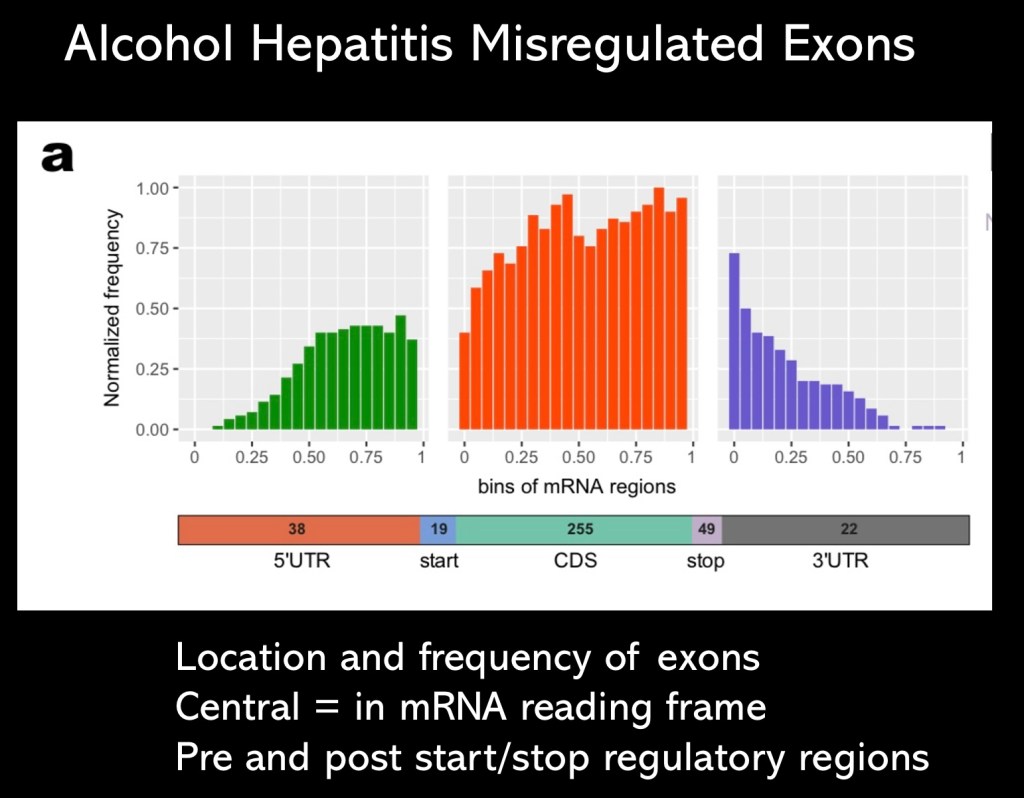
Histogram demonstrating distribution of misregulated exons occuring in acute hepatitis arranged by location. Exons within the coding frame are shown centrally in red. Data highlights the large number of misplaced exons contributing to disease progression and lack of expected repair and regeneration.
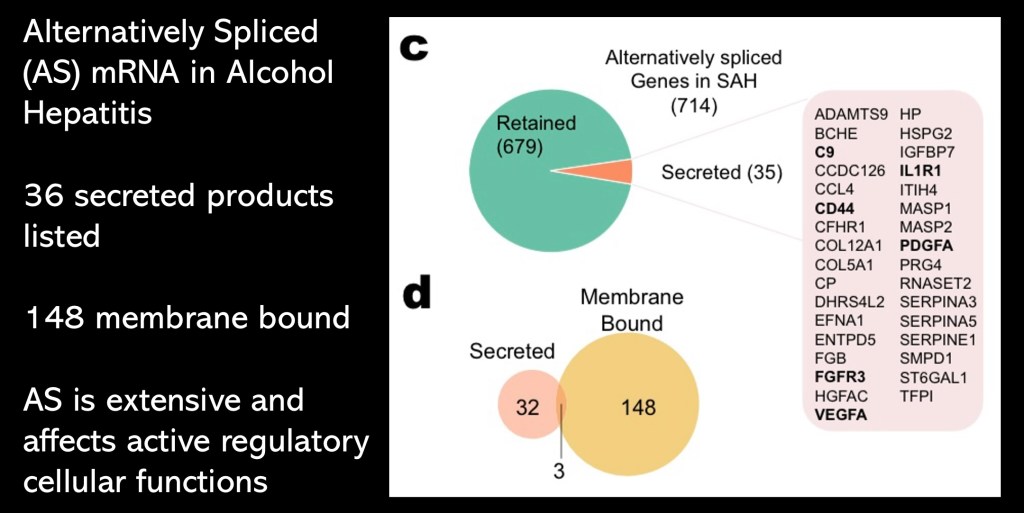
Cellular RNA sequence analysis found 714 alternative splices. 35 of these resulted in secreted proteins active in cell functions. The specific proteins are listed in the table above.
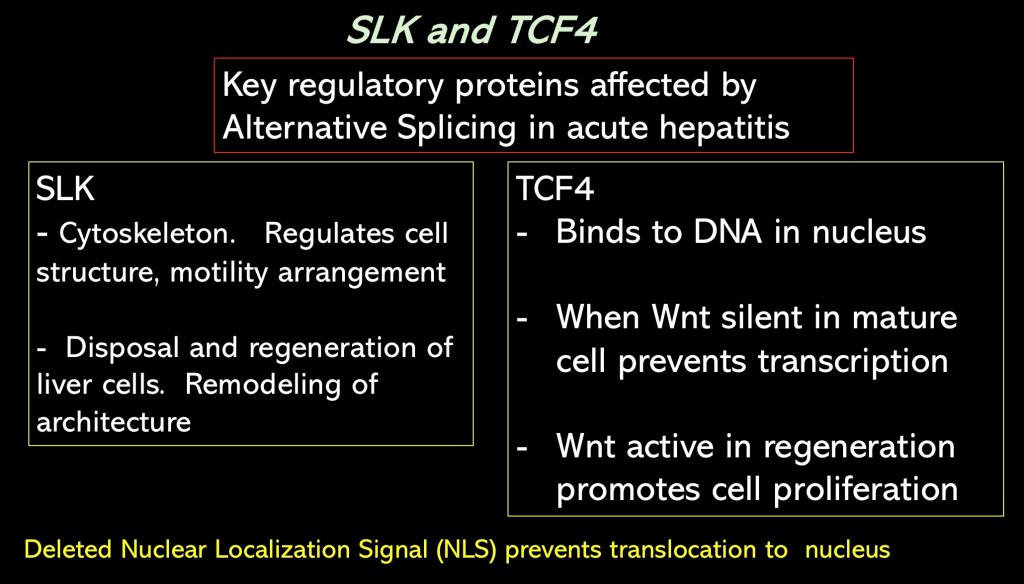
The authors focused on alternative splicing affecting these two regulatory proteins.
SLK is involved in regulation of the cytoskeleton and architectural arrangement of liver cells.
TCF 4 is a regulator of DNA transcription. It is a downstream effector in the Wnt pathway. When Wnt is silent in mature cells TCF 4 prevents gene transcription. When Wnt is activated TCF 4 results in transcription and initiates hepatic regeneration and proliferation of new hepatocytes.
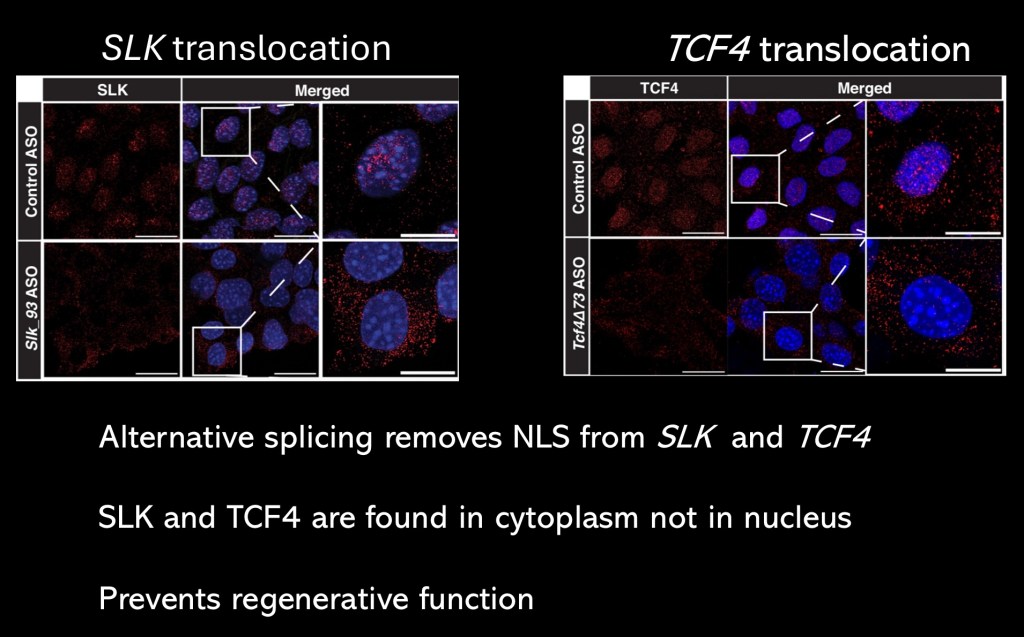
Fluorescent markers labeling SLK and TCF 4. Normal controls are at the top rows. SAH hepatocytes are in the bottom rows. In both cases there is no activity present within the cell nucleus. Neither of these transcription factors are then able to bind to nuclear DNA and carryout their normal functions.
Without cytoskeletal and architectural instructions from SLK cells cannot form functional tissue. Without TCF 4 hepatocytes do not receive signals to begin the regenerative process.
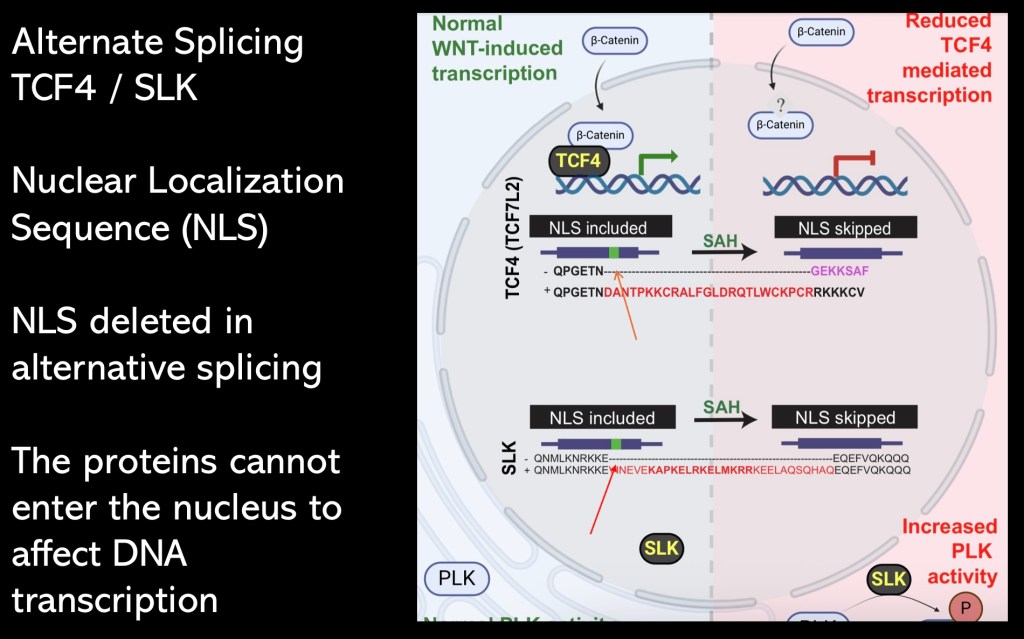
Protein translation takes place in the cytoplasm. In order to alter DNA transcription the protein must cross into the nucleus. The RNAs coding for transcription factors contain a nuclear localization sequence (NLS). This sequence will configure the protein in a way enabling translocation into the nucleus.
Using single nucleus sequencing the investigators found that in SLK and TCF 4 the NLS had been deleted by alternative splicing.
A substantial number of cases of severe alcohol liver disease continue to progress despite alcohol abstinence. The molecular mechanisms responsible have been unknown. This study used an in depth approach to identify pathways underlying biological substrates using a number of techniques including RNA sequencing at the single cell level.
Composition of cell types and inflammatory markers along with oxidative stress reveal a milieu whereby normal regulatory processes are disrupted.
This results in a large number of altered intracellular protein levels including RNA binding proteins.
RNA alternative splicing is compromised in these cases with a large number of misregulated exons present. The study focuses on marked under expression of the ERSP2 binding protein and the Wnt regenerative signaling pathway.
Due to splicing errors SLK and TCF4 fail to translocate into the cell nucleus. Thus, expression of genes required for orchestration of tissue components and transition to new functional hepatocytes is significantly impaired.
There are currently no specific treatment options for severe acute liver disease other than transplantation. Identifying specific molecular processes involved may allow for development of effective pharmacotherapy.

For information and educational purposes only. Images and data obtained from sources freely available on the World Wide Web. This list should not be considered medical or professional advice.
Comments and suggestions are always welcome jeffk972261@gmail.com
Jk 10/25
REFERENCES
Liver RNA
Dysregulated RNA splicing impairs regeneration in alcohol-associated liver disease
· Ullas V. Chembazhi, Sushant Bangru, Rajesh Kumar Dutta,
Chembazhi, U.V., Bangru, S., Dutta, R.K. et al. Dysregulated RNA splicing impairs regeneration in alcohol-associated liver disease. Nat Commun 16, 8049 (2025). https://doi.org/10.1038/s41467-025-63251-2
https://www.nature.com/articles/s41467-025-63251-2#citeas
,……………………………………………….
…………………………………………………
………………………………………………..
,snRNA
https://www.sciencedirect.com/science/article/pii/S2589004224028554
…………………………………………………
ESRP
https://www.sciencedirect.com/science/article/pii/S2589004222014778
…………………………………………………
TGF-β
https://www.nature.com/articles/nrneph.2016.48
………………………………………………….
WNT pathway
Progenitor cell expansion and impaired hepatocyte regeneration
in explanted livers from alcoholic hepatitis
Laurent Dubuquoy, Alexandre Louvet
Gut. 2015 December ; 64(12): 1949–1960. doi:10.1136/gutjnl-2014-308410.
………………………………………………………..
Why do we use steroids, Maddrey’s Discriminant Function, and the Lille score in Alcohol-Associated Hepatitis?
January 9, 2022
…………………………………………………………….
https://www.antibodies.com/applications/western-blotting
Western blot
……………………………………………………………..
https://cellandbioscience.biomedcentral.com/articles/10.1186/s13578-017-0153-7
NMD
…………………………………………………………….


Leave a comment